Abstract
Background: ABL kinase domain (KD) mutations cause resistance to tyrosine kinase inhibitors (TKIs) in chronic myeloid leukemia (CML) patients through impaired binding of TKI to the target site. To date, several mutations have been strongly associated with clinical resistance and have different susceptibility to TKIs depending on the type of mutation. One of the characteristics of patients with ABL KD mutation is the fact that some patients have multiple mutations and show poor prognosis for treatment than patients with single mutation. Although multiple mutations can be firstly detected by Sanger sequencing, it is not possible to determine whether mutations are in the same clone (compound mutations) or in different clones.
Aims: The aim of this study was to investigate the composition and pattern of mutations in the individual clones with serial samples from the patients carrying multiple mutations by subcloning sequencing.
Methods: Between 2002 and 2015, 735 CML patients had been screened for mutation analysis due to suspicious resistance to various TKIs including imatinib, nilotinib, dasatinib, bosutinib, radotinib, ponatinib and ABL001 using Sanger sequencing. Among them, 45 patients had multiple mutations. We performed Sanger sequencing and subcloning sequencing using 218 peripheral blood samples from the patients. For subcloning sequencing, at the least 20 colonies were selected at random and sequenced from all samples except 3 samples, and in total 4357 colonies from 218 serial samples are analyzed.
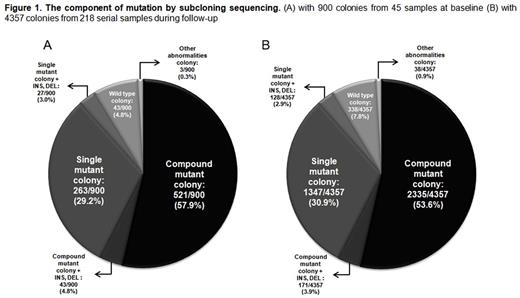
Results: The component of mutation by subcloning sequencing is shown in Figure 1. The disease status at the first multiple mutation detection (baseline) were in the frontline imatinib resistance (n=11), 2nd line TKI-resistance (n=29), and more than 3rd line TKI-resistance (n=5).
By Sanger sequencing with 45 baseline samples, 19 major mutations were detected and the higher frequency of mutation was in order T315I (29; 64.4%), E255K (15; 33.3%), Y253H (7; 15.6%), G250E (6; 13.3%), and F317L (5; 11.1%). Among 900 colonies with subcloning sequencing, the compound mutation was found in 564 (62.7%) colonies. And the colonies with single mutation, wild type sequence and other abnormalities (base insertion or deletion) were in 290 (32.2%), 43 (4.8%) and 3 (0.3%) respectively. A total of 552 mutant types were detected and the higher frequencies were in order T315I (324 colonies; 36.0%), E255K (132 colonies; 14.7%), Y253H (63 colonies; 7.0%), G250E (60 colonies; 6.7%), M351T (59 colonies; 6.6%) and E255V (48colonies; 5.3%).
In additional analysis with 4357 colonies of 218 serial samples on further treatment, 2506 colonies (57.5%) had compound mutation and the composition of non-compound mutation was also similar with that of baseline samples. Of 2506 colonies with compound mutation, a majority of colonies had double (53.4%) or triple (28.7%) mutations. The T315I and P-loop mutations were in 29.4% and 28.5% respectively. Known ABL001 resistant mutations were detected in 65 clones (1.5%).
Conclusion: The majority of multiple mutations were in compound mutations, and T315I and P-loop mutation were the major component of multiple mutation. As some minor clones with known TKI resistant mutations can be selected with further TKI treatment, these results highlight the need to identify the complexity of mutation using more sensitive techniques.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal